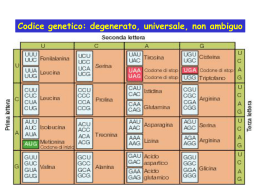
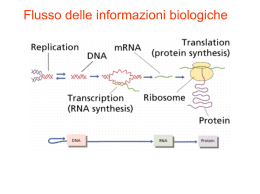
Inizio dell’RNAi • dsRNA (80-100 nt) in piante, C. elegans, Drosophila • siRNA in mammiferi Figure 2 Dicer and RISC (RNA-induced silencing complex). a, RNAi is initiated by the Dicer enzyme (two Dicer molecules with five domains each are shown), which processes double-stranded RNA into ~22-nucleotide small interfering RNAs. Based upon the known mechanisms for the RNase III family of enzymes, Dicer is thought to work as a dimeric enzyme. Cleavage into precisely sized fragments is determined by the fact that one of the active sites in each Dicer protein is defective (indicated by an asterisk), shifting the periodicity of cleavage from ~9–11 nucleotides for bacterial RNase III to ~22 nucleotides for Dicer family members. The siRNAs are incorporated into a multicomponent nuclease, RISC (green). Recent reports suggest that RISC must be activated from a latent form, containing a double-stranded siRNA to an active form, RISC*, by unwinding of siRNAs. RISC* then uses the unwound siRNA as a guide to substrate selection31. b, Diagrammatic representation of Dicer binding and cleaving dsRNA (for clarity, not all the Dicer domains are shown, and the two separate Dicer molecules are coloured differently). Deviations from the consensus RNase III active site in the second RNase III domain inactivate the central catalytic sites, resulting in cleavage at 22-nucleotide intervals. Meccanismo 1. Produzione siRNA 2. Formazione complesso RISC 3. Attivazione complesso RISC 4. Taglio dell’RNA bersaglio Figure 1 Double-stranded RNA can be introduced experimentally to silence target genes of interest. In plants, silencing can be triggered, for example, by engineered RNA viruses or by inverted repeat transgenes. In worms, silencing can be triggered by injection or feeding of dsRNA. In both of these systems, silencing is systemic and spreads throughout the organism. a, A silencing signal moves from the veins into leaf tissue. Green is green fluorescent protein (GFP) fluorescence and red is chlorophyll fluorescence that is seen upon silencing of the GFP transgene. b, C. elegansengineered to express GFP in nuclei. Animals on the right have been treated with a control dsRNA, whereas those on the left have been exposed to GFP dsRNA. Some neuronal nuclei remain florescent, correlating with low expression of a protein required for systemic RNAi. c, HeLa cells treated with an ORC6 siRNA and stained for tubulin (green) and DNA (red). Depletion of ORC6 results in accumulation of multinucleated cells. Stable silencing can also be induced by expression of dsRNA as hairpins or snap-back RNAs. d, Adult Drosophilaexpress a hairpin homologous to the white gene (left), which results in unpigmented eyes compared with wild type (right). Caratteristiche dell’RNAi • Può agire a diversi livelli: trascrizionale, posttrascrizionale, traduzionale • Può amplificarsi e diffondersi nell’organismo • Può coinvolgere sequenze adiacenti Viral Suppressors of RNAi The genomes of three positive-strand RNA viruses that encode suppressors of RNAi (black boxes) are shown. The polymerase genes (white boxes), other viral proteins (gray boxes), viral protease cleavage sites (diamonds), and sgRNA promoters (bent arrows) are indicated for comparison. Controllo di acidi nucleici “parassiti” • C. elegans senza RNAi hanno una aumentata mobilità di trasposoni endogeni • In alcuni sistemi i trasposoni “silenziati” si trovano organizzati in eterocromatina Regolazione dell’espressione genica • Mutazioni in componenti dell’RNAi causano alterazioni dello sviluppo (Arabidopsis, C. elegans, Drosophila) • Regolazione del gene Stellate in Drosophila Ruolo biologico dell’RNAi • Difesa dai virus (nelle piante VISG) • Controllo dei trasposoni • Regolazione dell’espressione genica Uso dell’RNAi • Studio sistematico della funzione dei geni • Terapia genica Fig. 1. The DNA vector-based RNA interference (RNAi) technology. (a) Generation of a hairpin siRNA directed by a Pol III promoter. An inverted repeat is inserted at the ©≠1 position of the U6 promoter (2351 to ©≠1). The individual motif is 19–29 nt, corresponding to the coding region of the gene of interest. The two motifs that form the inverted repeat are separated by a spacer of three to nine nt. The transcriptional termination signal of five Ts are added at the 30 end of the inverted repeat. The resulting RNA is predicted to fold back to form a hairpin dsRNA as shown. The resulting siRNA starts with either a G or an A at the 50 end, dependent on the promoter used (U6 or H1) and ends with one to four uridines, forming a 30 overhang that is not complementary to the target sequences. (b) Generation of two complementary siRNA strands synthesized by two U6 promoters. Two U6 promoters either placed in tandem or on two separate plasmids (not shown) direct transcription of a sense and an antisense strand of 19-nt RNAs. The two RNA strands are predicted to form a duplex siRNA in the transfected cells, with 30 overhangs of one to four uridines. Small temporal RNA (stRNA) Micro RNA (miRNA) • Piccole molecole di RNA (20-22 nt) • Prodotte da precursori di 90-100 nt trascritti autonomamente (anche policistronici) o maturati da introni • Si legano a mRNA che hanno sequenze complementari • Presenti in tutti gli eucarioti Nancy Standart et al. Genes Dev. 2007; 21: 1975-1982 Kosik Nature Reviews Neuroscience 7, 911–920 (December 2006) | doi:10.1038/nrn2037 Figure 5 A model for the mechanism of RNAi. Silencing triggers in the form of doublestranded RNA may be presented in the cell as synthetic RNAs, replicating viruses or may be transcribed from nuclear genes. These are recognized and processed into small interfering RNAs by Dicer. The duplex siRNAs are passed to RISC (RNA-induced silencing complex), and the complex becomes activated by unwinding of the duplex. Activated RISC complexes can regulate gene expression at many levels. Almost certainly, such complexes act by promoting RNA degradation and translational inhibition. However, similar complexes probably also target chromatin remodelling. Amplification of the silencing signal in plants may be accomplished by siRNAs priming RNAdirected RNA polymerase (RdRP)-dependent synthesis of new dsRNA. This could be accomplished by RISC-mediated delivery of an RdRP or by incorporation of the siRNA into a distinct, RdRP-containing complex. Kosik Nature Reviews Neuroscience 7, 911–920 (December 2006) | doi:10.1038/nrn2037 Scheme for the function of different snmRNAs by targeting bacterial or eukaryal mRNAs or pre-mRNAs leading to regulation of gene expression. Binding of a snoRNA to the splice-site of a eukaryal pre-mRNA or involvement in mRNA editing and modification has recently been proposed as one of several alternatives for a functional role, but remains to be experimentally verified. CARATTERISTICHE DI UN "SISTEMA VIVENTE" replicazione evoluzione DOGMA CENTRALE DNA RNA proteine CAPACITA' DIVERSE DELL'RNA RNA come genoma: virus a singolo o doppio filamento RNA come catalizzatore: meccanismo basato su cationi bivalenti (metalloribozimi) Apparecchio di Miller (1953) DIFFICOLTA' DELLE SINTESI PREBIOTICHE rese basse ribosio instabile (isomeri) legame glicosidico a o b fosfato solo al 3' e 5' legami fosfodiesterici solo 5'-3' FUNZIONI DELL'RNA NELLA CELLULA MODERNA traduzione rRNA, mRNA, tRNA maturazione rRNA snoRNA, RNasi MRP splicing snRNA, introni gruppi I e II maturazione tRNA RNasi P sintesi DNA primers, telomerasi traslocaz. proteine srpRNA PERCHE' L'RNA? l'RNA deve essere venuto prima del DNA l'RNA deve essere venuto prima delle proteine molti coenzimi hanno un ribonucleotide nella struttura l'RNA può agire come catalizzatore EVOLUZIONE SINTESI PROTEICA 1) tRNA 2) tRNA sintetasi 3) protoribosoma 4) codice genetico 5) mRNA 6) proteine PASSAGGIO RNA DNA 1) sintesi dei precursori del DNA 2) trascrizione inversa 3) replicazione 4) trascrizione SELEX (SYSTEMATIC EVOLUTION OF LIGANDS BY EXPONENTIAL ENRICHMENT) es. affinity column
Scaricare