PCR competitiva e Real Time
Competitive PCR
Competitive PCR
Homologous Competitor
Target mRNA
Competitor
Heterologous Competitor
Competitor
Target mRNA
Standard PCR using test primers and known amounts of target DNA (35 cycles of amplification).
Real time PCR
CT = cycle at which the observed fluorescence is 10-fold above background (10x amplification).
Real Time PCR
Real-Time PCR
• Precise detection of PCR products during
synthesis.
• Useful for:
– Genotyping
– Gene expression
• Real-time PCR is the standard for confirming
miroarray results
– Mutant detection
SYBR Green I Dye Chemistry
Fluorescence
Detection
95°
72°
55°
Taq-Man Probes
Assays can be multiplexed
TaqMan - Real-Time PCR
Specificity of Real-Time PCR
• For SYBR-Green, specificity is same as conventional
PCR.
– Disassociation analysis can be used to identify
“primer-dimers”.
• For Taq-Man, specificity is determined by specific probe
hybridization so specificity is very high.
Real Time QUANTITATIVE PCR
(quando si fa anche una curva standard)
Detection of Single Nucleotide Polymorphisms
Using RT-PCR
LUX
Primers are oligonucleotides labeled with a single fluorophore, custom-synthesized according to the
DNA/RNA of interest. Typically 20-30 bases in length, they are designed with a fluorophore close to the 3’ end in a
hairpin structure. This configuration intrinsically renders fluorescence quenching capability (c’e’ una G al 5’!!!); no
separate quenching moiety is needed. When the primer becomes incorporated into double-stranded PCR product, the
fluorophore is dequenched, resulting in a significant increase in fluorescent signal. This signal increase is the basis for
the LUX™ detection platform.
•Detect 100 or fewer copies of target genes
•Dynamic range of seven orders of magnitude
•Melting curve analysis
•Easy multiplexing
Hybridization Probes
Fluorescence
Detection
95°
72°
55°
Assays can be
multiplexed
Molecular beacons
At the annealing temperature, molecular beacons also bind to
the amplicons, undergo conformational reaorganization, and
generate fluorescence. When the temperature is raised to allow
primer extension, the molecular beacons dissociate from their
targets and do not interfere with polymerization. A new
hybridization takes place in the annealing step of every cycle,
and the intensity of the resulting fluorescence indicates the
amount of accumulated amplicon
SCORPION PROBES
Scorpions probes are highly sensitive, sequence-specific, bi-labeled fluorescent probe/primer
hybrids designed for Real Time PCR.
The Scorpions probe consists of a single-stranded bi-labeled fluorescent probe sequence held
in a hairpin-loop conformation with a 5' end reporter dye and an internal quencher dye directly
linked to the 5' end of a PCR primer via a blocker. The blocker prevents the polymerase from
extending the PCR primer.
At the beginning of the PCR reaction, the polymerase extends the PCR primer and
synthesizes the complementary strand of the specific target sequence. During the next cycle,
the hairpin-loop unfolds and the loop-region of the probe hybridizes intramolecularly to the
newly synthesized target sequence. Now that the reporter dye is no longer in close proximity to
the quencher dye, fluorescence emission may take place. The fluorescent signal is detected by
the qPCR instrument and is directly proportional to the amount of target DNA.
http://www.premierbiosoft.com/tech_notes/Scorpion.html
SCORPION PROBES
Figure 1. Scorpion probing mechanism. Step 1: initial denaturation of
target and Scorpion stem sequence. Step 2: annealing of Scorpion
primer to target. Step 3: extension of Scorpion primer produces doublestranded DNA. Step 4: denaturation of double-stranded DNA produced in
step 3. This gives a single-stranded target molecule with the Scorpion
primer attached. Step 5: on cooling, the Scorpion probe sequence binds
to its target in an intramolecular manner. This is favoured over the
intermolecular binding of the complementary target strand.
Digital PCR
A sample is diluted and partitioned into
hundreds or even millions of separate PCR
reaction chambers so that each contains one
or no copies of the sequence of interest. By
counting the number of 'positive' partitions (in
which the sequence is detected) versus
'negative' partitions (in which it is not),
scientists can determine exactly how many
copies of a DNA molecule were in the original
sample.
QuantaLife’s droplet digital PCR (ddPCR) technology converts a DNA sample into 20,000 1 nL droplets.
TaqMan-based amplification takes place in each droplet, followed by absolute quantitation of the number of
copies of a gene target as the individual droplets stream past a fluorescence detector.
Lo studio del trascrittoma
1
61
121
181
241
301
361
421
481
541
601
661
721
781
841
901
961
1021
1081
1141
1201
1261
1321
1381
1441
1501
GAATTCACGC TTTAAGGCTA
TAAATAAATA ACCAATACAA
ACAGCAAATC TCTTCTTCAC
GAGCTGAAGA CATAAATTGG
CATCATTTAT TTCACTGATG
ATTCTACAGT ACACAGTGAA
TAAACTCACA ACAGTATATC
TCTGTTTAAC CTGCATGATC
CTAGTTTTGA
ATATAATCAT
Gene 5
Gene 4
CAAAATGGGA
GACTATGCAA
Gene 3
CAATTATGGC
TTTGCTTTAC
Gene 2
GGTATTCCTA CCACAACCTT
Gene 1
AAGGTTGTTC TATACCCAGA
TACATATTTT TCTTACTTCT
GATTTTTTTT TTCCTTATGA
TGGGAGTGGT GGCTCATGCT
CCTGAGGTCA GCAGTTACAG
AATACAAAAA TTTGCCAGGT
GAGGCAGGAG AATCGCTTGA
TTGGACTCTA GCAGGGTGAC
TTTCCCATAT GAAAAAAATA
AGGTCATAGA TGTAATCTTT
AACATATTCC ATGCCGTCAG
GCTACCTCAA GGATAAGAAG
GGGGCTGCTC CTTCCTGGGC
GAGATGAATG CCCATGTTTT
TGGCCACCTT TAAATAAAGT
ATCTTTTCAG AGACAAATGC
CTGTTTGCTT GTTTAGAGTT
TAACCAATGG AATTATCTGG
TGCAAATTTA TTCCGTACCA
Genoma
TCATGTATAC TTAGTCAAGT
AGCTCATGAT GGGTAAATGA
TCACTCTTTA GTATTTGCTT
GATATGGAGA GACAAGTGAA
ATGCTGAAAT GAGAATTAAT
TTTTTGCCCG TAAGAGACAT
GCTTAGCATA GTGGTTGACT
TTTCAACGTG ATTGCTATGC
CACTTTCTTT TTCTTCTTGG
TCTCAAGAAA TTTTTCTCAT
TGTAATCCCA GCACTTTGGG
ATGAGCCCGG CCAAAATGGT
GTGGTGGCAG GCACTTGTAA
ACCCAGGAGG CAGAGGTTGC
AAGAGCAAAA CTCCATCTCA
ACACAAGATC CGGAATACAG
CTTCCAGGAA AAATTTATTT
ATAGCACTGG CTTAGGAGAC
CAGGCAAAAG GCAAGCACAG
AAGTTTCAGA AACTCACTGA
CCCGAAGGGA GAACTGATGC
ACAGCATTAC TAAAAAAAAA
ATTCTCTGAC ATCTGAGGTT
GTAATATTTG CTTTGGTGTA
CCTCAGACTT TATTTATTTT
GCAAATGTCA ATTTAATTAT
TGTAAATACA CTAAACCATA
CTTTTCCCTG AGAAAGAGTA
CTTTAGTCGA CGTTTGTTTC
ATCACCACAA
TTTTGTTTTC
Gene 10
ACATCCAAAA
TATCGAACCA
Gene 9
Gene 8
GTGGCCTAGA
ATAGGTGGCA
Gene 7
AAATATAAAT
TTTAGAGATG
Gene 6
CCACTTCACT TTCTTTAAAA
TTGACATTTT TTGGCTCAGG
TGAAAAAGAC ATAATCGTGC
AGGCTGAGGC TGGTGGATCA
GAAACCTCAT CTCTACTAAA
TCCCAGCCAC TCGGGAGGCT
AGTGAGCCAA GATCATTCCA
GGAAAAAAAA AATCATAAAT
AGAGGAGCAT AATCCTTTGC
CAGATAAGAC CAGAATTGGA
GAATGAGGAG GAGCCTGCAG
GGGCGGCATG CACTCACACT
CAGAGCTAGC AGCTCCCATA
TTAGAAAGGC TGAATGACTT
L’espressione orchestrata del corredo genico è alla base
del mistero della vita
cDNA
Avian myelobastosis virus (AMV) reverse transcriptase was the first RTase specifically purified for use in first strand cDNA reactions. The active enzyme
consists of two subunits that together encode the DNA polymerizing activity, and an RNase activity called RNase H which degrades RNA in RNA:DNA
heteroduplexes. The RTase of Moloney murine leukemia virus (MMLV) is a single polypeptide chain that encodes all the required RTase functions. The
MMLV RTase has been cloned and re-engineered to have negligible levels of RNase H activity, without compromising its first strand cDNA polymerizing
function
Sintesi cDNA doppio filamento
Librerie cDNA
Rapid Amplification of cDNA Ends (RACE)
New RACE
Figure 2. RLM-RACE for Mouse CXCR4 Gene and
Xenopus TGF-ß Related Gene. Total RNA from mouse
liver and Xenopus embryos (stage 41) were analyzed
using Ambion's RLM-RACE Kit. Initial PCR (A) and
nested PCR (B) products were analyzed on a 2%
agarose gel with EtBr staining. The results indicate that
a product of the expected size is amplified after nested
PCR. Note that CXCR4 is a moderately expressed
message while the TGF-ß related gene is an extremely
rare message.
SMART RACE (Clontech)
Low Throughput Analysis of Gene
Expression
– Northern blot hybridization
– RT-PCR
– Quantitative real-time PCR
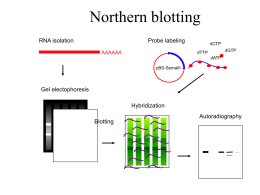
Northern Blot Hybridization
Formaldehyde agarose gel
and northern blot (GAPDHprobed) of poly A+ mRNA
isolated from total RNA from
various samples.
RT-PCR Assay
Reverse transcription was
oligo-dT primed and PCR
carried out using actinspecific primers. A: Poly A+
mRNA was isolated from
0.25, 0.5, or 1 µg mouseliver total RNA (lanes 1 - 3).
C: negative control. B: mRNA
was isolated from 103 or 104
HeLa cells (lanes 1 & 2),. M:
123 bp ladder.
RT Real-Time PCR
• Can reliably detect 2-fold differences
in initial template concentration.
• Standard curve required for absolute
quantitation
• Requires some sort of reference reaction to
normalize template input.
• Endogenous gene sequence
• 18s ribosomal RNA
Mid Throughput Analysis of Gene
Expression
– Reverse northern
– EST
– Differential libraries
Reverse Northern
• Filtri di nylon sui quali sono stati trasferiti i cloni
corrispondenti ai geni di cui si vuole monitorare
l’espressione, vengono ibridati con sonde ottenute dalla
marcatura radioattiva della popolazione di cDNA
• Permette di monitorare il livello di espressione genica di
molti geni in parallelo (Es.: cloni isolati in esperimenti
Differential Display o cDNA-AFLP)
Expressed Sequence Tags (ESTs)
ESTs are short (200–500 nucleotides) DNA sequences that can be used to identify a
gene that is being expressed in a cell at a particular time.
The Procedure:
• Isolate the mRNA from a particular tissue (e.g., liver)
• Treat it with reverse transcriptase. Reverse transcriptase is a DNA
polymerase that uses RNA as its template. Thus it is able to make genetic
information flow in the reverse (RNA ->DNA) of its normal direction (DNA ->
RNA).
• This produces complementary DNA (cDNA). Note that cDNA differs from the
normal gene in lacking the intron sequences.
• Sequence 200–500 nucleotides mainly at the 5′ end of each cDNA.
• Examine the database of the organism's genome to find a matching
sequence.
• That is the gene that was expressed.
Electronic Northern
• Le Expressed Sequence Tags sono
sequenze di 500 bp ottenute dal
sequenziamento random di una libreria di
cDNA
• Le frequenze delle EST corrispondenti ad
un gene forniscono una misura del livello
di espressione di quel gene in una data
libreria
Librerie sottrattive
(obsolete)
Una libreria sottrattiva di cDNA contiene i cDNA ottenuti dagli
mRNA prodotti in determinate condizioni/tessuti/organi (target) ma
non in altre (driver)
E’ ottenuta mediante sottrazione degli RNA del driver dagli RNA del
target
Differential Screening
Screening con
sonde a cDNA del
tipo cellulare A
Poco sensibile verso trascritti poco rappresentati (< 0.1%)
Screening con
sonde a cDNA del
tipo cellulare B
Libreria sottrattiva a cDNA
Una libreria sottrattiva contiene i cloni di cDNA corrispondenti agli mRNA
presenti in una cellula o in un tipo cellulare e non in un secondo tipo.
Target
Driver
RsaI
AluI
EcoRI
Miscelazione con un eccesso di 50
volte del cDNA driver, riscaldamento
a 95 °C e annealing
Ligazione alle braccia di λgt10
• Sensibile (mRNA < 0.001%)
• Può confrontare solo 2 campioni alla volta
• Il confronto è “monodirezionale”
PCR-Select
Tecnologie per l’analisi su ampia scala dei
profili di espressione
• Metodi basati sulla visualizzazione di profili di
frammenti di cDNA su gel ad alta risoluzione:
– Differential Display
– cDNA-AFLP (AFLP-TP)
• Metodi basati sul sequenziamento dei trascritti:
– SAGE (LongSAGE, SuperSAGE)
– RNA-Seq
• Metodi basati sull’ibridazione di cDNA marcato a sonde
immobilizzate su un supporto solido:
– Macroarray
– Microarray
Differential Display
(obsoleto)
Principio:
• Retrotrascrizione e amplificazione delle
estremità 3’ degli mRNA con un set di primer
oligo(dT)12 ancorati e un decamero arbitrario.
Combina:
• Potenza della PCR
• Elevata risoluzione di una separazione
elettroforetica su gel di poliacrilammide
denaturante
Retrotrascrizione
con un primer
oligo(dT)12 degenerato
5’-TTTTTTTTTTMN-3’
M = G, A, C
N = G, A, T, C
Amplificazione
Separazione
Identificazione e
Isolamento delle
bande
Caratteristiche principali
• Sensibile
• Non necessita della
conoscenza delle
sequenze analizzate
• Permette di
confrontare più
campioni
contemporaneamente
• Scarsa riproducibilità
tra diversi laboratori
• Produce falsi positivi
• Difficile da rendere
quantitativa
cDNA-AFLP
Deriva dalla tecnica AFLP per la generazione di marcatori molecolari
Amplified
Fragment
Length
Polymorphisms
Si basa sulla retrotrascrizione degli mRNA, il taglio con enzimi di
restrizione dei cDNA prodotti e la loro ligazione ad adattatori di
sequenza nota. I frammenti così generati vengono amplificati con
primer specifici disegnati sulla sequenza degli adattatori.
Combina:
• Potenza della PCR
• Elevata risoluzione di una separazione elettroforetica su gel di
poliacrilammide
• Specificità della reazione di taglio enzimatico e di condizioni di PCR
ad elevata stringenza
Retrotrascrizione
Digestione con esacutter
Digestione con tetracutter
Ligazione degli adattatori
Amplificazione
Separazione elettroforetica dei frammenti
Separazione elettroforetica dei campioni e rilevazione
per autoradiografia
A
123
B
123
C
D
E
F
G
H
I
123
123
123
123
123
123
123
Caratteristiche
• Non necessita della conoscenza a priori delle sequenze
dei trascritti analizzati
• Le condizioni di elevata stringenza della reazione di
PCR hanno permesso di risolvere i problemi di bassa
riproducibilità osservati con il Differential Display
• Può essere ottimizzato in modo da diventare una tecnica
quantitativa (primer in conc. limitante)
• Il frammento generato non è strettamente all’estremità 3’
e quindi è più informativo nel caso di sequenze ignote
• Produzione di falsi positivi
• Frammenti MseI-MseI BstYI-BstYI
• Alcuni mRNA potrebbero non essere rappresentati
(mancanza del sito di taglio o in posizione errata)
Serial Analysis of Gene Expression
(SAGE)
• Si basa sulla generazione di brevi (14 bp)
frammenti di cDNA derivanti dalla
retrotrascrizione degli mRNA che vengono
poi concatenati e sequenziati.
• Permette l’analisi dei profili di espressione
di migliaia di trascritti contemporaneamente
AAAAA
TTTTT
mRNA
Oligo(dT) biotinilati
Retrotrascrizione a cDNA
Taglio con un anchoring enzyme (AE) NlaIII e legame con
sferette magnetiche coniugate a streptavidina
Suddivisione in 2 porzioni
e legame a 2 linker
Taglio con tagging enzyme (TE)
BsmFI e fill-in con Klenow
Ligazione e amplificazione
con i primer A e B
Taglio con anchoring enzyme (AE) NlaIII
XXXXXXXXXXOOOOOOOOOOCATG
GTACXXXXXXXXXXOOOOOOOOOO
Isolamento dei ditag, concatenazione
e clonaggio
• Non sempre l’enzima utilizzato produce tag delle
stesse dimensioni
• Una lunghezza di 15 bp può non garantire la
specificità del tag: per questo motivo è stata
sviluppato il protocollo LongSAGE che utilizza
l’enzima MmeI che permette di produrre
tag di 21 bp
• L’efficienza di ligazione varia con le ultime basi
adiacenti all’estremità piatta del tag
• Non tutti i trascritti contengono la sequenza
dell’enzima selezionato e non sono quindi inclusi
nell’analisi
SuperSAGE
• Utilizza l’enzima di tipo III EcoP15l che
permette la produzione di sequenze tag
> 25 bp
• Consente l’identificazione univoca dei
trascritti consentendo una accurata analisi
di espressione
Se fosse possibile monitorare l’espressione dell’intera batteria genica presente in una
cellula o in un tessuto, si potrebbe:
• costruire un “catalogo genetico” dei processi biologici che avvengono in quelle
cellule
• comprendere il set di geni necessari per quei processi
E comparando i “profili di espressione genica” di due diversi tipi cellulari, si potrebbe
capire cosa rende quelle cellule diverse da ciascun’altra.
Microarrays
La rapida individuazione delle “pagine importanti del libretto
di istruzioni” accelera in modo davvero formidabile lo studio
dei geni “chiave”
Genetic content of microarrays
Nucleic acid
sequences
• cDNA
• Genomic DNA
• Oligonucleotide
Spots – features targets
Microarray tecnology
• cDNA spotting
Many Companies - Custom array spotters
• Oligo spotting
Operon – MWG - Custom array spotters
• Oligo synthesis on chip
Affymetrix – NimbleGen - Agilent - Combimatrix
cDNA microarray
• Libraries of cDNA clones,
• expressed sequence tags
(EST),
PCR Products
• clones isolated from
subtraction libraries
• ORF in genomic DNA
• cDNA-AFLP analysis
cDNA microarray
(300–800 nucleotides)
Universal
primers
bacterial cultures,
purified plasmids,
cDNA, RNA
(Ethanol precipitation – Membrane purification)
Oligonucleotide microarrays
(20–70 nucleotides)
Disadvantages over cDNA targets:
•Knowledge of sequences required
Benefits over cDNA targets:
•Different parts of the same gene can be represented on the array
• Discriminate between related gene sequences and study different
members of gene families simultaneously
• Oligonucleotide targets are readily available from commercial
manufacturers or synthesized by researchers
• Less time and effort required to prepare oligonucleotides
Production methods of microarray
Non-contact deposition
Production methods of microarray
Contact deposition
Comparison of characteristics of
different microarray printing methods
Contact pen printing
Microtiter
plate well
volume
(microliters)
Uptake
volume
(microliters)
Spot volume
(nanoliters)
Spot size
(micrometers)
Piezoelectric printing
Syringe-Solenoid printing
10–30
20 – 50
20 – 50
0.2 – 1.0
5 – 10
5 – 10
0.5 – 2.5
5 – 100
0.1 – 10
75 – 250
250 – 500
125 – 175
Slide surface chemistries
Aminosilane coated slides
Unmodified DNA can be attached to amine-modified slides, via interactions between negatively charged phosphate
groups on the DNA and the positively charged slide surface. This interaction helps ensure denaturation of the DNA
as well as increase its binding affinity to the slide surface. UV treatment can be used to further immobilize the DNA onto
the slide surface. Attachment via electrostatic interactions is suitable for binding DNA fragments that are longer than
60–70 nucleotides. For attaching oligonucleotides to aminemodified glass, chemical coupling methods must be
used.
Slide surface chemistries
Aldehyde coated slides
Amino-modified DNA can be attached to microarray slides that have been
modified with aldehyde groups . The amino group can be introduced into DNA in a
PCR amplification reaction using aminomodified oligonucleotides.
Limitazioni dei microarray prodotti per
“deposizione”
• Limitato numero di geni che
possono essere analizzati
simultaneamente (<10,000)
• Elevata probabilità di errore
umano
• Limitata uniformità dei segnali
osservati
• Scarsa flessibilità
Microarray prodotti con oligonucleotidi
sintetizzati in situ
NimbleGen
Affymetrix
•
•
•
•
Quartz chip coated with a light-sensitive chemical compound
Lytographic masks are used to transmit light onto specific locations of
the wafer
The surface is flooded with a solution containing a nucleotide (A or T or
G or C) and coupling occours only on light activated regions
The coupled nucleotide also contain a light sensitive group, so the cycle
can be repeated till the completion of the probes synthesis
GeneChip by Affymetrix
Typical density: 1.6
cm up to 400 000
features
20 x 4 = 80 masks
$2000/mask
Affymetrix: features
Affymetrix probes are 25 nucletides long
The most recent Affymetrix human genome array has more than 1.3 million
“features” (11um) with an array density of 106 probes / cm2
Affymetrix: probe sets
Affymetrix uses a probe set of 11-16 probes per gene. Each probe set consist
of pair of probes, each one formed by one probe with perfect match (PM)
and one probe with one central nucleotide mismatch (MM). This allows
to evaluate signal due to cross-hybridization
Affymetrix Array
16 perfect
match oligos
16 mismatch
oligos
Affymetrix
•
•
Highly processive (industrial scale)
Large user base
•
•
•
Static, non flexible design
Needs a specific reader
Very sensitive to single mismatches
APPLICATIONS
•
•
•
•
•
Whole Genome: Linkage analysis, association studies, population genetics,
chromosome copy number
Targeted Genotyping: SNPs
Expression analysis
Expression regulation analysis
Human, plant, bacteria chips available
NimbleGen
NimbleGen’s Maskless Array
Synthesis (MAS) technology combines
photodeposition chemistry with
digital light projection to shorten
array fabbrication from months to
hours
• Extreme flexibility
• Highly reproducible array
fabrication and statistically
robust results
• Length up to 70 mer
385,000 to 2.1 million unique probe
features are synthesized in a single array
1plex : 462€
4plex : 508€
HD2 1plex : 1200€
Custom design fee : 1000€, waived if you order more or 10 arrays
Agilent
• Agilent uses HP’s inkjet printing technology to deliver
nucleotides
• Oligos are sinthesyzed base-per-base in repetitive print layers
using standard phosphoramidite chemistry
Evolution of Printing Technology
150 µm
Recent scans of 30µm features on
prototype scanner based on
existing instrument
115 µm
65 µm
30 µm
22K
44K
244K
1M
2002
2004
2006
2008
Sufficient 10 µm scanning resolution
Needs 5 µm scanning resolution
Needs 2 µm scanning resolution!
5um
2um
1M
244k
2006
105k
44k
15k
2007
2008
400k
180k
60k
Agilent
• 60-mer oligo format yield more sensitivity
compared to 25-mer format.
• Expensive masks are not required: flexible
design
• Standard chip format (can be used with
standard microarray scanners)
CombiMatrix
Silicon
wafer
Silicon
Microarray
• Software applies voltage
to sets of specific
electrodes
• Electrode activation
controls chemical
reactions at each
individual electrode on the
microarray
BUFFER
Signal In
H+
e-
Combimatrix: synthesis process
START
Combimatrix: synthesis process
ELECTROCHEMICALLY
DETRITYLATE
(Deprotect)
Combimatrix: synthesis process
COUPLE
Combimatrix: synthesis process
WASH AND REPEAT PROCESS WITH
SEQUENTIAL AMIDITE EXPOSURE
Combimatrix: synthesis process
Deblock
Combimatrix: synthesis process
Couple
Combimatrix: synthesis process
Wash
CustomArray™ Formats
12K
4x2K
2x40K
90K
Features and Benefits:
Configuration: 1 array having
up to 90,000 programable sites
for custom probe synthesis.
Hyb Chamber is disposable and
comes free of charge.
Open architecture
Re-usable: Strip and re-hyb
up to 4 times after 1st usage.
• whole transcriptome chips
• dedicated chips
• microRNA chips
Sample labelling for gene expression analysis
Use of controls in microarray experiments
Negative controls
Positive controls
Labelled DNA
Housekeeping gene
controls
DNA sequences that should not hybridize
with any labelled probe
Verifying that the target DNA is binding
effectively to the slide surface during the
hybridization and washes
Actin, glyceraldehyde 3-phosphate
dehydrogenase (GAPDH), and tubulins
Microarray scanning
Number of replicates required for microarray
platforms
based on Affymetrix chip (Jorstad et al. 2007 Trends in Plant Science)
•
At the statistical level: 3 replicates is the strict minimum.
•
With microarray it depends on 2 parameters:
•
–
The threshold of false positives differentially expressed genes (FDR): The maximum is 10% for a global
screening but good analysis require a strict maximum of 5% and often less (1%). At least 5 replicates are
required for a maximum of 1%.
–
The threshold of truly differentially expressed genes discovered (TDR): To detect the maximum of
differentially expressed genes, more than 20 replicates are required (used in cancer research for example).
To detect half of the differentially expressed genes, 3 technical replicates are enough.
Those 2 parameters are linked each other:
–
Global screening projects to identify highest differentially expressed genes could require a minimum of 3
replicates (with high FDR and/or low TDR).
–
Projects to identify the maximum number of differentially expressed genes require a strict minimum of 8
replicates (TDR=80%) and can reach 20-30 replicates for high precision (TDR>90%).
–
Projects to identify a large part of genes differentially expressed (TDR=40-70%) require a minimum of 3
replicates (FDR=5%) to 5 replicates (FDR=1%) to recover half of the genes differentially expressed.
The number of replicates required depends also on good experiment
design (no dilution of the signal) and good technical hybridization
Applications
Genomic analysis:
SNP detection and analysis
(Affymetrix).
25-base oligos. 4 oligos for SNP:
CCGTGACGGGAGCTGATGATCGTGT
CCGTGACGGGACCTGATGATCGTGT
CCGTGACGGGAACTGATGATCGTGT
CCGTGACGGGATCTGATGATCGTGT
OLIGO
CCGTGACGGGAGCTGATGATCGTGT
perfect match
...GGCACTGCCCTCGACTACTAGCACA...
High signal
CCGTGACGGGAGCTGATGATCGTGT
...GGCACTGCCCTCGACTACTAGCACA...
Lower signal
SAMPLE
mismatch
one base difference in the length of the oligo is enough to substantially
diminish hybridization signal
Applications
Genomic analysis:
SNP detection and analysis with APEX
1. Up to 6000 known 25-mer oligos
are immobilized via 5’ end on a
coated glass surface (‘DNA
chip’).
2. Complementary fragment of 3. Template dependent single
PCR amplified sample DNA is nucleotide extension by DNA
annealed to oligos.
polymerase. Terminator
nucleotides are labeled with 4
different fluorescent dyes.
4. DNA fragments
and unused dye
terminators are
washed off.
Signal detection.
Applications
Gene expression analysis
Applications
Gene expression analysis
There are far fewer protein-coding genes in the human genome than there are proteins
in the human proteome (~22,000 genes vs. ~400,000 proteins).
22,000 genes
Vs.
400,000 proteins
Applications
Gene expression analysis
The large increase in protein diversity is thought to be due to alternative
splicing and post-translational modification of proteins.
There may be many undiscovered multiple-exon protein-coding genes
By the use of an exon microarray tens of thousands of potential new exons have
been discovered in mouse
Comparative Genomic Hybridization (CGH)
The Power of Comparative Genomic Hybridization
• Establish correlations between gene copy number and disease biology
• Screen multiple genomic targets for copy changes
• Detect single copy changes with unparalleled sensitivity
• Accelerate the development of products for genomic disease management to guide therapeutic interventions
Comparative Genomic Hybridization (CGH)
Comparative genomic hybridization is a molecular genetic method for analysing copy
number variations (CNVs) relative to ploidy level in the DNA of a test sample
compared to a reference sample.
CGH is only able to detect unbalanced chromosomal abnormalities. This is because
balanced chromosomal abnormalities such as reciprocal translocations, inversions or
ring chromosomes do not affect copy number, which is what is detected by CGH
technologies. CGH does, however, allow for the exploration of all 46 human
chromosomes in single test and the discovery of deletions and duplications, even on
the micro scale which may lead to the identification of small duplications or deletions
in candidate genes. Indeed, through the use of dedicated DNA microarrays it is
possible to measure CNV with increased resolution as low as a few bases.
Human Whole-Genome aCGH
With a capacity of 385,000 isothermal probes, NimbleGen's high-density, long oligo arrays span the entire nonrepetitive regions of the human genome in a single array, tiling the entire genome at a median probe spacing of
6,000bp. NimbleGen's Human Whole-Genome aCGH, available as a catalog design, is a cost-effective platform
for genome-wide analysis of copy number changes. Unlike other commercial whole-genome aCGH platforms,
NimbleGen's Human Whole-Genome aCGH tiles through both genic and intergenic regions of the genome,
providing the most thorough, unbiased coverage available.
Figure A: Human Whole-Genome aCGH
Genome-wide chromosomal gains and losses were detected in a human cancer cell line
by whole-genome aCGH. Data are processed with a 60kb averaging window and
displayed as a log2 ratio plot.
Figure B: Focused Fine-tiling Validation
A region of copy number variation in chromosome 6 identified by whole-genome aCGH is confirmed by fine-tiling
aCGH using a high-resolution chromosome 6 microarray (400bp median probe spacing). A zoom view of an
amplified region (indicated by the blue box) depicts the distal breakpoint of the copy number gain and shows
additional copy number variants ranging in size from 6kb to 70kb. Annotation tracks showing known transcripts
and regions of normal copy number variation are also displayed in the zoom-in.
Custom Fine-Tiling aCGH
NimbleGen's Custom Fine-Tiling aCGH can
be
used
to
detect
deletions
and
amplifications and to map the associated
breakpoints with unprecedented resolution.
You can choose your region(s) of
interest for a fine-tiling array design
with probes spaced as dense as
10bp. This density of probe placement
enables ultra-high resolution mapping of
breakpoints to an interval that can be
validated by PCR amplification and
sequencing. The selected regions need not
be contiguous or even within the same
chromosome. For example, the copy number
changes identified in a whole-genome aCGH
experiment can be combined and tiled at
higher resolution on a Custom Fine-Tiling
CGH Array.
Resolution of Cytogenetic
Technologies
Technique
Resolution (kb)
Detectable Alterations Include
Karyotype
10,000-100,000
Large amplifications, deletions, insertions;
translocations; interband inversions
Conventional CGH
3,000-10,000
Large amplifications, deletions, insertions; chromosomal
copy number changes
SKY/M-FISH
500-1,500
Large & small amplifications, deletions, and insertions;
translocations; copy number changes
FISH
50-100
Large & small amplifications, deletions, insertions,
translocations, and chromosomal copy number changes
minimally involving the 1-2 genes found within the
FISH probe fragments
Array-based CGH
using BACs
100-200
Large and small amplification, deletions, and insertions
Array-based CGH
using cDNAs
0.5-2
Individual gene amplifications, deletions and insertions
Array-based CGG
using oligos
0.06 - 0.3
Individual gene amplifications, deletions and insertions
Sequencing
0.001 (1 bp)
SNPs, small insertions and deletions
ChIP on chip
ChIP on chip
ChIP-on-chip (also known as ChIP-chip) is a technique that combines chromatin
immunoprecipitation ("ChIP") with microarray technology ("chip"). Like regular ChIP,
ChIP-on-chip is used to investigate interactions between proteins and DNA in vivo.
The goal of ChIP-on-chip is to locate protein binding sites that may help identify
functional elements in the genome. For example, in the case of a transcription factor
as a protein of interest, one can determine its transcription factor binding sites
throughout the genome. Other proteins allow the identification of promoter regions,
enhancers, repressors and silencing elements, insulators, boundary elements, and
sequences that control DNA replication. If histones are subject of interest, it is
believed that the distribution of modifications and their localizations may offer new
insights into the mechanisms of regulation.
WHAT IS ChIP on chip
(or Location Analysis) ?
Chromatine Immunoprecipitation Analysis identifies
where proteins bind to DNA to regulate transcription.
There are less than 2000 TF and around 25000 human genes. 1 TF can
interact with several genes. But WHAT TF for WHAT Genes ??
ChIP on chip help map the binding sites and answer this question
Promoter region
Transcription Factor
Gene
Chromatin immunoprecipitation (ChIP)
Epigenetics
The study of heritable changes in gene expression or function
that occur without changes in the DNA sequence itself
DNA methylation is the only known epigenetic modification of DNA in
mammals
Agilent Confidential
Page 139
June
Page2007
139
DNA methylation
Gene regulation
– embryonic development
– genomic imprinting
– gene silencing - cancer
Methylation of C5 of cytosine in CG dinucleotide
– DNA methyltransferases
– Post-replication maintenance (DNMT 1)
– de novo (DNMT3A & DNMT3B)
CpG islands
– regions of high CG, generally un-methylated, 1% of human genome
– promoter associated
Chromatin stability
Agilent Confidential
Page 140
June
Page2007
140
Scarica