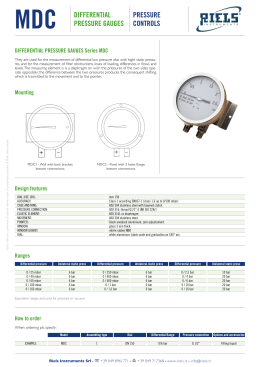
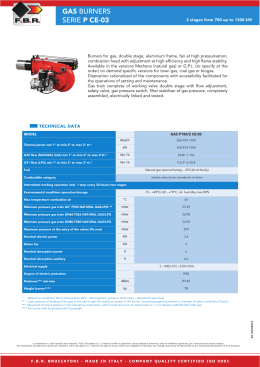
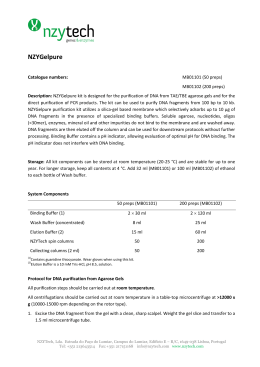
Energy Driven Undocking of FKBP12 Ligands Using Hamiltonian Replica Exchange Simulations Francesca Nerattini and Piero Procacci Dipartimento di Chimica, Università di Firenze January 29, 2014 Methods The Absolute binding free energy for drug-receptor complexes are determined using Hamiltonian Replica Exchange (H-REM). In the extended multicanonical system, bound and unbound states are sampled using a H-REM scaling protocol whereby the drug-receptor interactions are progressively weakened with increasing replica index. A weak tethering potential prevents the drug to drift away in the bulk solvent. The absolute free energy of binding is computed as ∆G = −kB T ln(pb /pu ) − kb T ln(Veff /V0) where V0 and Veff are the standard volume and the mean accessible volume of the ligand imposed by tethering potential. The probability for the bound and unbound state, pb , pu are evaluated using the full Generalized Ensemble statistic exploiting the Multiple Bennett acceptance Ratio re-weighting technique, i.e. N X pb = pu N X wi (xi )H(F xi )) / i wi (xi )[1 − H(F (xi ))] i with H being an Heaviside step function discriminating bound and unbound states based on appropriately defined contact function F and w are the MBAR weights. The algorithm Schematic representation of HREM simulation of a tight binding ligand (in red) with solute-solvent counter-scaling. Each horizontal line represents a replica simulation in the generalized ensemble with scaled active-site-drug potential function. The thickness of the line for each replica is proportional to the MBAR weight of the corresponding GE configurations. Unbound states are found only in the last replicas with a small MBAR weight. The system(s) Six FKBP12 complexes have been simulated: SB3-FKBP12(wt) and -FKBP12(Ile56Asp); N-Elte-FKBP12(wt) and -FKBP12(Ile56Asp); FK506-Elte-FKBP12(wt) and -FKBP12(Ile56Asp); Results Figure: Extrapolation of the binding free energy as a function of the number of contacts of the FK506 natural drug (left) and of the new synthetic N-Elte compound (right) vs native FKBP12 and the mutant I56D-FKBP12 Simulation Details H-REM Molecular Dynamics simulation of solvated FKBP12 with the ligands sb3, N-Elte and the Tacrolimous natural drug lasting from 4 to 10 ns launched on 16 replicas. All atom Amber03 force field, 15000 atoms in total, multiple time step simulation (0.5 to 12. fs steps) with Particle Mesh Ewald for electrostatics. Figure: Residues contribution to the binding for FK506 (left) and N-Elte (right)
Scarica