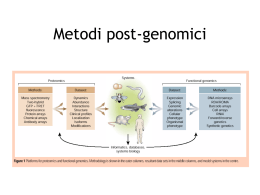
Metodi post-genomici in biochimica cellulare Metodi post-genomici Metodi post-genomici Quantitative analysis of systems biology by taking advantage of available genomic information at the level of 1. 2. 3. 4. 5. 6. 7. SNPs analysis associated to disease or drug response mRNA (transcriptomics) Protein (proteomics) Post-translational modifications (aka “modificomics”) Surface exposure (surfomics) Protein-protein interactions (interactomics) Small metabolites (metabolomics) and their relations (metabonomics) Many other fantasy exercises (glycomics, lipidomics, allergenomics, degradomics, excluding – perhaps – comics...) G.B Smejkal, “I’m an –omics, you’re an -omics... ” Exp. Rev. Proteomics 3 (2006) 383-385 Metodi post-genomici • • • • Modelli cellulari e animali Trascrittomica Proteomica Systems biology Modelli cellulari e animali (farmacologici e/o genetici) Modelli cellulari (farmacologici e/o genetici) • Facilità di mantenimento e trattamento • Possibilità di combinare trattamento farmacologico e manipolazione genetica • Utili per riprodurre un singolo meccanismo • Cellule umane (o murine) Modelli animali (farmacologici) • Intero organismo vs. cellule isolate • Trattamento sistemico o lesione chimica locale • Possibilità di valutare l’effetto anterogrado/retrogrado Modelli animali (e vegetali?!?) (genetici) • • • • • Organismi modello Genoma noto Non solo topo! Vita breve Invertebrati (e piante…) Trascrittomica Trascrittomica Trascrittomica Trascrittomica Trascrittomica • Distanza Euclidea • Correlazione di Pearson Proteomica • Non c’è correlazione tra quantità di mRNA e quantità di proteina (Gygi et al., 1999) • Il proteoma è un’istantanea del fenotipo a livello biochimico • Il proteoma tiene conto del processing delle proteine S.P.Gygi et al., Mol. Cell. Biol. 19, 1720 (1999) Proteomica • Metodi basati su 2-DE • Metodi “gel-free” Two-dimensional electrophoresis (2-DE) Staining pI 3-10 NL 170 kDa 95 kDa 72 kDa 170 kDa 95 kDa 72 kDa 55 kDa 43 kDa 55 kDa 43 kDa 34 kDa 34 kDa 26 kDa 26 kDa 17 kDa 17 kDa Group B Group A pI 3-10 NL Proteomica (2-DE) Vantaggi • Possibilità di caricare campioni non purificati • Risoluzione estremamente alta • I gel 2 –DE sono collettori di frazioni proteiche molto efficienti • Proteine sono protette all’interno della matrice del gel Problematiche • Gradiente di pH • Limiti nel determinare proteine poco rappresentate • Capacità di caricare campione • Proteine idrofobiche • Proteine ad alto peso molecolare Proteomica (2-DE) • A global, unbiased approach • Hypothesis-generating rather than hypothesis-driven • A “find the difference” game between two conditions Control Treated Proteomica (2-DE) Proteomica (2-DE) Proteine Colorazione Acquisizione Analisi di immagine Proteomica (2-DE) Find the difference… Controllo Esordio Precoce Esordio Tardivo Proteomica (2-DE) Find the difference… Proteomica (2-DE) Metodi statistici Proteomica (2-DE) Identificazione delle proteine • Peptide mass fingerprinting • LC-MS/MS • Western blot (non globale) Proteomica (2-DE) Peptide Mass Fingerprinting Proteomica (2-DE) Peptide Mass Fingerprinting Proteomica (2-DE) Peptide Mass Fingerprinting (Limiti) • La proteina non è presente nel database • La proteina è ricca di modificazioni co/posttraduzionali • Lo spot nasconde più di una proteina Proteomica (2-DE) Peptide Mass Fingerprinting (Limiti) • La proteina non è presente nel database • La proteina è ricca di modificazioni co/posttraduzionali • Lo spot nasconde più di una proteina Proteomica (2-DE) LC-MS/MS Differential in-gel electrophoresis (DIGE) Matching not needed o High cost Spatially accurate o Weak signal Sensitive to small quantitative changes o Only binary comparison DIGE Control [Cy5] Pharmacological Treatment [Cy3] Gel-based vs. Gel-free Webb-Robertson and Cannon, Brief Bioinform 2007;8:304-317. Poor detection of acidic- basic- proteins poor solubility of membrane proteins limited loading capacity of gradient pH strips (crowding effect) Low reproducibility of gels relatively low throughput 2D gels perform robust separations 2D gels are well-suited for PTM analysis Parallel, quantitative and label-free readout Monteoliva and Albar, BRIEFINGS IN FUNCTIONAL GENOMICS AND PROTEOMICS. VOL 3. NO 3. 220–239. Proteins do the job, not peptides Proteomica (gel-free) • Metodi quantitativi (ICAT, iTRAQ, …) • Protein arrays Gel-free MS & Proteomics Quantitative Proteomics • Labelling (ICAT, iTRAQ, SILAC, 18O enrichment, …) • Label free (AQUA, SRM/MRM, …) Isotope-coded affinity tagging (ICAT) Isobaric Tagging for Relative and Absolute Quantitation (iTRAQ) Selected/Multiple reaction monitoring (SRM/MRM) Proteomica (gel-free) • Protein arrays (e SELDI) What next? • You will call your preferred MS expert to ask her/him to identify your spots • You will get a list of protein names • What tells you that list? Systems Biology Systems Biology • Necessità di analizzare un elevato numero di informazioni (Network analysis) • Necessità di arricchire un ridotto numero di informazioni (Network enrichment) Systems Biology • Interazione fisica • Stesso pathway (KEGG) • Stessa Gene Ontology (GO) Protein Networks Cellular processes are regulated by protein interaction networks Protein networks: • control development programs • regulate signal transduction pathways • manage metabolic pathways • are based on physical interactions or cellular localization Protein networks Graph: a graphical representation of elements (nodes) connected by edges. Nodes are proteins, edges are interactions Protein networks Hub: connecting several nodes Subnetwork Protein networks Regulating interactions: Controls Inhibits Feedback Interacts with… Building protein networks •Co-occurrence in databases •Physical interactions •Genomic proximity •Expression •Proteomics •Literature (pubmed) •Pathways (KEGG, Reactome, …) •GO Terms Available Databases • Free, online PPI data – IntACT (EBI) http://www.ebi.ac.uk/intact/ – DIP http://dip.doe‐mbi.ucla.edu/dip/Main.cgi – MINT http://mint.bio.uniroma2.it/mint/Welcome.do – BIND/BOND http://bond.unleashedinformatics.com/ – HPID http://wilab.inha.ac.kr/hpid/ – UniProt http://www.uniprot.org/ – NCBI Entrez Gene http://www.ncbi.nlm.nih.gov/sites/entrez?db=gene • Pathways – Reactome http://www.reactome.org/ – KEGG http://www.genome.jp/kegg/pathway.html – Panther http://www.pantherdb.org/pathway/ – NCI Nature PathwayInteractionDb http://pid.nci.nih.gov/ – BioPATH http://www.molecular‐networks.com/biopath/index.html • Commercial applications – GeneGO, Ingenuity Pathway Analysis… GO enrichment Physical Interaction- IntAct Physical Interaction- IntAct Physical Interaction- IntAct KEGG Pathways, Reactome String 9.0 http://string-db.org/ String 9.0 BioProfiling http://www.bioprofiling.de/ BioProfiling Global networks R spider R spider implements the Global Network statistical framework to analyze gene list using as reference knowledge a global gene network constructed by combining signaling and metabolic pathways from Reactome and KEGG databases. Reactome is an expert-authored, peerreviewed knowledgebase of human reactions and pathways. Reactome database model specifies protein-protein interaction pairs. The meaning of "interaction" is broad: 2 protein sequences occur in the same complex or they occur in the same or neighbouring reaction(s). Both, Reactome signaling network and KEGG metabolic network were united into the integral network. For the human genome, the resulting integral network covers about 4000 genes involved in approximately 50,000 unique pairwise gene interactions. Reference: if you will find the results produced by R spider usefull, please cite: 1. Antonov A.V., Schmidt E., Dietmann S., Krestyaninova M.,Hermjakob H. R spider: a network-based analysis of gene lists by combining signaling and metabolic pathways from Reactome and KEGG databases Nucleic Acids Research, 2010, Vol. 38, No. suppl_2 W118W123 PPI spider PPI spider implements Global Network statistical framework to analyze gene/protein list using as reference knowledge a global protein-protein interaction network from IntAct database. For the human genome, the reference network covers about 7960 genes involved in approximately 40,000 unique pairwise interactions. Reference: if you will find the results produced by PPI spider usefull, please cite: 1. Antonov A.V., Dietmann S., Rodchenkov I., Mewes H.W. PPI spider: A tool for the interpretation of proteomics data in the context of protein protein interaction networks. PROTEOMICS. Volume 9, Issue 10, 10 May 2009. ProfCom What is Cytoscape? http://www.cytoscape.org/ What is Cytoscape? “Cytoscape is an open source bioinformatics software platform for visualizing molecular interaction networks and integrating these interactions with gene expression profiles and other state data” Fully customizable through plugins http://www.cytoscape.org/ Cytoscape plugins BiNGO (Biological Network Gene Ontology) Tool per determinare le GO statisticamente sovrarappresentate Consensus PathDB ConsensusPathDB-human integrates functional interaction networks including complex protein-protein, metabolic, signaling and gene regulatory interaction networks in Homo sapiens. Data originate from currently 24 public resources for functional interactions, interactions that we have curated from literature. Additionally, biochemical pathways have been imported from several databases for use in pathway analyses. Data are integrated in a complementary manner and redundancies are avoided. MCODE MCODE is a Cytoscape plugin that finds clusters (highly interconnected regions) in a network. Clusters mean different things in different types of networks. For instance, clusters in a protein-protein interaction network are often protein complexes and parts of pathways, while clusters in a protein similarity network represent protein families. Un esempio (realizzato a Busto…) Un modello cellulare per identificare nuovi meccanismi e nuovi bersagli terapeutici Quali condizioni? Il Modello Un nuovo bersaglio! Trova le differenze! Cosa hanno in comune? 71 Il modello cellulare • La linea cellulare umana SH-SY5Y incamera dopamina, ma la immagazzina con difficoltà nelle vescicole Simile a quello che succede nella malattia di Parkinson α-sinucleina • (Gómez-Santos et al., 2003) La linea viene trasfettata stabilmente per esprimere α-sinucleina or β-galattosidasi I livelli di α-sinucleina sono alterati nella malattia di Parkinson 72 Le condizioni sperimentali -Sinucleina Dopamina Controllo NON trattate Controllo trattate con Dopamina Effetto combinato α-Sinucleina NON trattate α-Sinucleina trattate con Dopamina 73 Trova la differenza! 74 Trova la differenza! Dopamina 11 α-Sinucleina 4 • sintesi proteica • citoscheletro • mitocondri • trascrizione • stress ossidativo • mitocondri • trasduzione del segnale Entrambi i fattori 8 75 Cosa hanno in comune? Dopamina α-Sinucleina Entrambi i fattori 76 Generare nuove ipotesi NF-κB Apoptosi 77 Ritorno al modello VDAC2 Ctr DA VDAC1 Ctr DA VDAC2 Ctr DA VDAC3 Ctr DA VDAC2 4h Inibitore GSK3β ctr ctr inib DA DA inib Alberio, Fasano, Rizzuto e altri, in preparazione 78 Partendo dal modello cellulare, verso nuovi bersagli terapeutici Alterazione della Dopamina Proteine che cambiano NF-κB, i VDACs, GSK3β. Nuove terapie? 79 Stage disponibili • Proteomica differenziale del ruolo dei mitocondri nella patogenesi della malattia di Parkinson • Validazione in larga scala di marcatori ematici di malattia di Parkinson
Scaricare