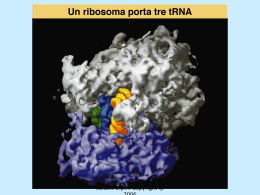
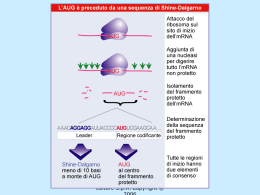
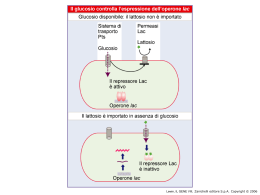
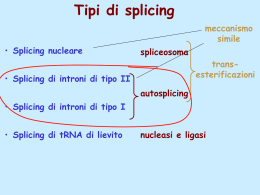
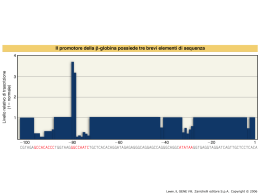
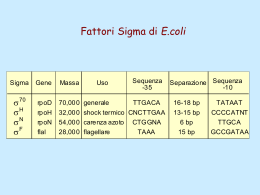
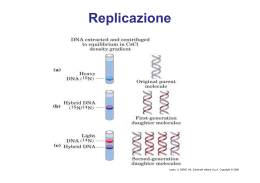
Dati sulla struttura del DNA • Contiene due catene • Le percentuali G=C e A=T • Legami fosfodiesterici tra nucleotidi Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 L’appaiamento delle basi implica la formazione di legami idrogeno H citosina N H N O N sugar H O N N H N CH3 timina H O H N sugar H N N H O N N N N adenina N sugar guanina N sugar Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Figura riportata nella pubblicazione originale di Watson e Crick Nature, Aprile 1953 20 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 La struttura del DNA (B) • Il DNA ha una forma ad elica regolare, diametro 20Å, passo 34Å • Legami idrogeno tra le basi • L’impilamento delle basi è determinato da interazioni idrofobiche • Ogni coppia è ruotata di 36° • Solchi maggiore (22Å) e minore (12Å) • Avvolgimento in senso orario (elica destrorsa) Nature , Vol. 171, p.737, April 25, 1953 MOLECULAR STRUCTURE OF NUCLEIC ACIDS A Structure for Deoxyribose Nucleic Acid We wish to s ugge st a structure for the salt of deoxyribose nucleic acid (D.N.A.). This structure has novel fea tures which are of considerable biological interest. A structure for nucleic acid has alr eady been proposed by Pauling and Corey (1). They kind ly made their manuscript availa ble to us in advance of publication. Their model consists of three intertwined chains, with the phosphates near the fibre axis, and the bases on the outside. In our opin ion, this structure is uns atisfactory for two reasons : (1) We believe that the material which gives the X-ray diagrams is the salt , not the free acid. Withou t the acidic hyd rogen atoms it is not cle ar what forces would hold the structure together, especia lly as the negatively charged phosph ates near the axis wil l repel each other. (2) Some of the van der Waals distances appear to be too small . Another three-chain structure has also been sugg ested by F raser (in the press). In his model the phosph ates are on the outside and the bases on the inside, li nked together by hyd rogen bonds. This structure as described is rather ill- defined, and for this reason w e shall not comm ent on i t. We wish to put forward a ra dically differ ent structure for the salt of deoxyribose nuc le ic acid. This structure has two helical chains each coile d round the same axis (see diagram). We have made the usual chemi cal assumptions, namely, that each chain consists of phosph ate die ster groups joining §-Ddeoxyribofuranose residues with 3',5' linkages. The two chains (but not their bases) are rela ted by a dyad perpend ic ular to the fibre axis. Both chains follow right - handed heli ces, but owing to the dyad the sequences of the atoms in t he two chains run in opposit e dire ctions. Each chain loosely resemble s Furberg's2 model No. 1; that is, the bases are on the inside of the heli x and the phospha tes on the outside. The configuration of the sugar and the atoms near it is close to Furberg's 'standard configu ration', the suga r being rough ly perpend icular to the attached base. There is a residue on each every 3.4 A. in the z-direction. We have assumed an ang le of 36΅ between adjacent residues in the same chain, so that the structure repeats after 10 residues on e ach chain, that is, aft er 34 A. The distance of a phosphorus atom from the fibre axis is 10 A. A s the phosphates are on the outside, cations have easy access to them. The structure is an open one, and its water content is rather high. A t lower water contents we would expect the bases to tilt so that the structure could become more compact. The novel feature of the structure is the manner in whic h the two chains are held together by the purine and pyrimi dine bases. The planes of the bases are perpend ic ular to the fibre axis. The are joined together in pairs, a single base from the other chain, so that the two lie side by side with identical z-co-ordinates. One of the pair must be a purine and the other a pyrimi dine for bonding to o ccur. The hyd rogen bonds are made as follows : purine posit ion 1 t o pyrimidine position 1 ; purine po sition 6 to py rim idine position 6. If it is assumed that the bases only occur in the structure in th e most plausible tautomeric forms (that is, wit h the keto rather than the enol configu rations) it is found that only sp ecifi c pairs of bases can bond together. These pairs are : adenin e (purine) with thym ine (pyrimi dine), and guan ine (purine) with cytosine (pyrimidine). In other words, if an adenine forms one me mber of a pair, on either chain, then on th ese assumptions the other me mber must be thymine ; si mil arly for guan ine and cytosine. The sequen ce of bases on a s ing le chain does not appear to be restric ted in any way. However, if only specifi c pair s of bases can be formed, it foll ows that if the sequence of bases on one chain is given, then the sequence on t he other chain is automatically determined. It has been found experimentall y (3,4) that the ratio of the amount s of adenine to thymi ne, and the ration of guanin e to cytosine, are always bery close to unity for deoxyribose nu cleic acid. It is probably impossible to build this structure wit h a ri bose sugar in place of the deoxyribose, as the extra oxyg en atom would make too close a van der Waals contact. The previously publ ished X-ray data (5,6) on d eoxyribose nucleic acid are in sufficient for a rigorous test of our structure. So far as we can tell, it is rough ly compatible with the experim ental data, but it must be regarded as unp roved unt il it has been checked against more exact results. Some of these are giv en in the foll owing comm uni cations. We were not aware of the detail s of the result s presented there when we devised our structure, which rests mainly though not entire ly on publi shed experime ntal data and stereochemical argume nts. It has not escaped our notice that the specific pairing we have postulated imm edia tely sugg ests a possible copying mechanism for the g enetic materia l. Full details of the structure, includ ing the condi tions assume d in buil ding it , together with a set of co-ordinates for the atoms, wil l be published elsewhere. We are much ind ebted to Dr. Jerry Donohue for constant advice and criticism, especia lly on interatomi c distances. We have also been stim ulated by a know le dge of the general nature of the unpub li shed experim ental results and ideas of Dr. M. H. F. Wilkins, Dr. R. E . Franklin and their co-workers at King's College, London. O ne of us (J. D. W .) has been aided by a fell owsh ip from the National Founda tion for Infantil e Paralysis. J. D. WATSON F. H. C. C RICK Medical Research Coun cil Unit for the Study of Mole cular Structure of Biological Systems, Cavendish Laboratory, Cambridge. 1. Pauling, L., and Corey, R. B., Nature, 171, 346 (1953); Proc. U.S. Nat. Acad. Sci., 39, 84 (1953). 2. Furberg, S., Acta Chem. Scand., 6, 634 (1952). 3. Ch argaff, E., for refere nces see Zamenho f, S., Brawerman, G., and Chargaff , E., Bio chim. et Biophy s. Acta, 9, 402 (1952). 4. Wyatt, G. R., J. Gen. Phys iol., 36, 201 (1952). 5. A stbury, W. T., Symp. Soc. Exp. Biol. 1, Nucleic Acid, 66 (Camb. Univ. P ress, 1947). 6. W ilkins, M. H . F., and Rand all, J. T., Biochim. et Biophys. A cta, 10, 192 (1953). It has not escaped our notice that the specific pairing we have postulated immediately suggests a possible copying mechanism for the genetic material. Il modello a doppia elica di Crick e Watson suggerisce un meccanismo di replicazione OLD OLD NEW NEW OLD NEW OLD NEW Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Strutture alternative del DNA Gli acidi nucleici possono formare vari tipi di doppia elica Gli acidi nucleici possono formare vari tipi di doppia elica Elica Coppie di basi per giro Rotazione tra due basi Diametro B 10 (10,4) 36 (34,6) 20 (19) A 11 32,7 23 Z 12 -30 18 Strutture superiori del DNA DNA circolare con superavvolgimento = 0 DNA superavvolto negativamente Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Il superavvolgimento negativo può convertirsi nella separazione locale dei due filamenti DNA superavvolto negativamente struttura Z (in corrispondenza di regioni di alternanza Pu/Py) separazione dei filamenti (denaturazione di regioni ricche in A+T) strutture cruciformi (in corrispondenza di palindromi) Superavvolgimento del DNA Positivo = DNA superspiralizzato Negativo = DNA sottospiralizzato Enzimi che alterano la topologia del DNA Topoisomerasi: tipo I tipo II (girasi) Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Enzimi per gli acidi nucleici • Polimerasi: DNA polimerasi RNA polimerasi • Nucleasi: esonucleasi endonucleasi Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006 Lewin, IL GENE VIII, Zanichelli editore S.p.A. Copyright © 2006
Scarica